Courtesy: GRDC Ground Cover, original article published online – Tomorrow’s fungicides: gene switches and nanobots | Groundcover (grdc.com.au)
New tools and techniques are being added to the fungicide resistance management arsenal.
Plant pathogens form intimate relationships with their plant hosts to gain access to resources for their growth and survival, often killing the plant host in the process.
These are complex relationships; the host and pathogen even undergoing genetic changes as they evolve together and with each trying to survive.
If additional selection pressure is placed on the pathogen in the form of a fungicide, applied to disrupt fungal growth and disease progression, a further level of genetic change can result – that of fungicide resistance.
This dynamic is often likened to an ‘arms race’ between the plant host and pathogen as both change at the genetic level to outmanoeuvre each other.
To help plants ‘outsmart’ these pathogens, researchers have increasingly sought to adopt more-sophisticated tools to keep ahead of the fungi and to understand the nuances of the plant-pathogen relationship.
Researchers at Curtin University’s Centre for Crop and Disease Management (CCDM) with GRDC support are at the forefront of fungicide resistance research.
They are working on two fronts to develop new detection methods for the molecular analysis of fungicide resistance, and to investigate the development of novel fungicides.
Identifying fungicide resistance at work
Fungal pathogens can develop resistance to fungicides in a range of ways, with some of these mechanisms being easier to identify than others. Small genetic changes (mutations) in the genes targeted by fungicides are one of the most common mechanisms that lead to resistance.
Another mechanism used by fungi is to ‘copy and paste’ the fungicide target gene in the genome. By gaining extra copies of the target gene, the fungus can overcome the inhibitory effects of fungicide by simply having more copies of the gene working within the cell, allowing it to outcompete the fungicide.
However, identifying when and how these gene ‘copy/paste’ events have happened in a fungal genome is difficult. It is hard to determine how frequently this form of fungicide resistance is occurring in a pathogen population and, consequently, how this affects the management of fungicide resistance.
Dr Chala Turo and Dr Kat Zulak are using long-read DNA sequencing technology that allows the sequencing of entire chromosomes. This has been pivotal to identifying ‘copy/paste’ events that have resulted in very high levels of resistance in fungal pathogens.
The ability to identify these gene duplications increases the ability to monitor pathogen populations for genetic changes. Such changes have already been identified in net blotch pathogen populations and can inform fungicide resistance management practices. Similar research is underway for powdery mildew.
Finding the master gene switch
Within every cell of your body is DNA which gives instructions to the cell on how to make proteins via genes. Each cell has the exact same DNA sequence, but it is the genes that are turned on or off that gives each cell its specific job.
The same is true for fungi. For example, some genes are only turned on when a spore germinates on a leaf and turned off again as the fungus grows. ‘Master regulator’ genes are important for deciding when whole batches of genes are switched on or off, much like how traffic lights regulate traffic flow.

Anjana Sharma is working to identify genetic ‘master switches’ in fungi to develop fungicides with new modes of action. Photo: CCDM
PhD candidate Anjana Sharma has identified a family of master regulator genes that are responsible for controlling dozens of downstream genes in the fungal pathogen Parastaganospora nodorum (the causal agent of Septoria nodorum blotch in wheat).
One by one, Ms Sharma is deleting each of these master regulator genes to identify the downstream network of genes that are controlled by these master switches. Ms Sharma is also seeing which of these master regulator genes are critical for the pathogen’s survival and ability to cause disease.
This research has the potential to reveal not just one gene, but whole suits of genes that fungi need to survive and cause disease.
Ms Sharma is hoping that future fungicides could be developed with new modes of action to target not only the master regulators but also the important genes that are downstream of the regulators and key to the survival of the pathogen.
Next-gen fungicides
Can we develop fungicides that are as effective as current chemistries, but have less impact on environmental and human health? This is the question that Dr Elizabeth Czislowski aims to answer with her research at CCDM.
It seems straight out of a sci-fi movie: nanoparticles that are made from folded pieces of DNA that are capable of catching spores before they land on a leaf, or which are programmed to weaken the cell wall of fungal pathogens.
Medical research developed DNA origami therapy for novel antiviral therapy. It is now being explored by CCDM as a new fungicide technology.
Medical research developed DNA origami therapy for novel antiviral therapy. It is now being explored by CCDM as a new fungicide technology: Photo image provided by J. Kretzmann, University of Western Australia.Reproduced from Sigl, C., Willner, E.M., Engelen, W. et al. Programmable icosahedral shell system for virus trapping. Nat. Mater. 20, 1281–1289 (2021). https://doi.org/10.1038/s41563-021-01020-4
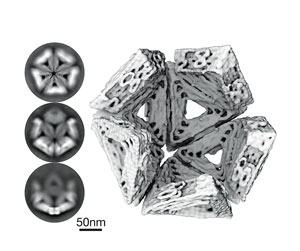
As futuristic as this sounds, it could well come to fruition as a future fungicide technology. While originally designed as an antiviral therapy, researchers at CCDM are exploring whether this nanoparticle technology could be adapted to fight fungal pathogens.
The technology offers many advantages including being biodegradable and having low or no toxicity to mammalian cells. Ongoing research at CCDM is working to understand the stability of the nanoparticles after coming into contact with fungi and how antifungal properties can be designed into the nanoparticles. Other future technologies being investigated include RNA-based biopesticides designed to specifically turn off fungal genes.
Future fungicides will have to strike a balance between technologies with potent antifungal activity and those with minimal effect on human or environmental health.
To ensure the future of crop production and the security of the world’s food supply these are the types that need to be harnessed to manage the continued threat of fungicide resistance.
More information: Associate Professor Fran Lopez-Ruiz, fran.lopezruiz@curtin.edu.au,

